 |
|
|
Sequence recognition for automatic vector removal with DNA Sequence Assembler
Content
1. General info about the pGEM-T eazy vector
Below we are showing the sequence of the pGEM(R)-T easy vector. The vector is delivered in a linear form, to facilitate the ligation of the insert. The pGEM(R)-T Easy Vector has been linearized with EcoRV at Base 60 of this sequence (indicated by an asterisk *) and a T added to both 3'-ends (the added T is not included in the sequence of the vector given below). Therefore, the insert will be ligated at position 60, where the asterisk * is found.
> pGEM-T Easy Vector (Promega corporation) 1 GGGCGAATTG GGCCCGACGT CGCATGCTCC CGGCCGCCAT GGCGGCCGCG
2. How to design recognition sequences
Step I: Identify the base positions where the insert will be ligated
In our example, it is position 60 (marked by an asterisk *):
> pGEM-T Easy Vector (Promega corporation) 1 GGGCGAATTG GGCCCGACGT CGCATGCTCC CGGCCGCCAT GGCGGCCGCG
Step II: add a T at the end of each flanking region (T marked in red) Because the T is added at the 3' end, before the asterisk (which is the 3' end of the given DNA strand) we add a T and after the asterisk (which is 5' end of the given DNA strand) we add an A, corresponding with a T at the 3' end on the complementary strand.
> pGEM-T Easy Vector (Promega corporation) 1 GGGCGAATTG GGCCCGACGT CGCATGCTCC CGGCCGCCAT GGCGGCCGCG
Step III: Identify the F recognition sequence by selecting 15-20 bases in the 1st flanking region (just before the asterisk, marked below in lime color)
> pGEM-T Easy Vector (Promega corporation) 1 GGGCGAATTG GGCCCGACGT CGCATGCTCC CGGCCGCCAT GGCGGCCGCG 51 GGAATTCGATT* AATCACTAGTG AATTCGCGGC CGCCTGCAGG TCGACCATAT 101 GGGAGAGCTC CCAACGCGTT GGATGCATAG CTTGAGTATT CTATAGTGTC 151 ACCTAAATAG CTTGGCGTAA TCATGGTCAT AGCTGTTTCC TGTGTGAAAT
The F recognition sequence is: GGCCGCGGGAATTCGATT
Step IV: Select 15-20 bases in the 2nd flanking region (just after the asterisk) (marked below in green)
> pGEM-T Easy Vector (Promega corporation) 1 GGGCGAATTG GGCCCGACGT CGCATGCTCC CGGCCGCCAT GGCGGCCGCG 51 GGAATTCGATT* AATCACTAGTG AATTCGCGGC CGCCTGCAGG TCGACCATAT 101 GGGAGAGCTC CCAACGCGTT GGATGCATAG CTTGAGTATT CTATAGTGTC 151 ACCTAAATAG CTTGGCGTAA TCATGGTCAT AGCTGTTTCC TGTGTGAAAT
The selected bases in the 2nd flanking region are: AATCACTAGTG AATTCGC.
Step V: Visually check that the F recognition sequence is different from the selected bases in the 2nd flanking region
The F recognition sequence is: GGCCGCGGGAATTCGATT If different, move to step VI. If not different, repeat steps III and IV, but this time select longer sequences.
Step VI: Obtain the R recognition sequence (marked in fuchsia) by making the reverse complement of the selected bases from the 2nd flanking region (step IV)
The selected bases in the 2nd flanking region are: AATCACTAGTG AATTCGC.
Step VII: Visually check that the F and the R recognition sequences are not identical
The F recognition sequence is: GGCCGCGGGAATTCGATT
3. How to input and use the recognition sequence with DNA Baser Assembler
Step I: Define your vectors
Click the Tasks button to open the 'Tasks' panel In the 'Tasks' panel, chose the desired task from ''Sequence processing' or 'Mutation detection' section
The PROJECT MANAGER window will open. Click the 'Vector Removal' tab:
In the Vector Removal tab you will be able to enter your vector recognition sequence(s). In the "Add new recognition sequence" box, enter the name and the nucleotides of the F recognition sequence file. Press the ADD button to add the sequence into the 'Current recognition sequences' list:
In the "Vector cleaning" box choose if you want to remove or to keep the recognition sequence when is found. The vector will be removed in both cases, just the recognition sequence will be kept if so is chosen by the user).
Repeat operation for the R recognition sequence. Make sure that both recognition sequences are active (the check box in front of them is checked): Press the APPLY button to save the settings.
Note: For details about vector removal and sequence recognition, click here to go to the Vector Removal page.
Step II: Sequence assembly
Click the PROJECT BUILDER tab, navigate to the folder containing your sequences and add them into the JOB LIST. Now you are ready to start the sequence assembly (or mutation detection) by pressing the "Start sequence assembly" button.
Step III: Contig inspection
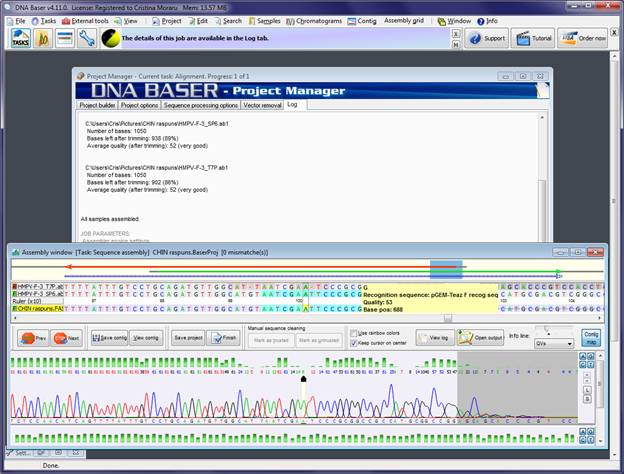
If the sequence assembly process was successful, the 'Assembly Window' will open. Here you can see if the recognition sequences were found and the vector was removed. The recognition sequences are marked in blue. The vector bases are strike. It will be automatically cut from your contig when you save the contig to disk.
In the screenshot above, the recognition sequence was not removed, as per user choice (see Step I). If you want it to be removed, you need to select the 'Cut recognition sequence' option in the 'Vector Removal' tab, BEFORE assembling the sequences.
In this example, both recognition sequences were found:
Custom support
If you need custom support with tasks similar to this one (sequence assembly, vector removal, primer design, automation of sequence cleaning/processing jobs, etc) we can provide it at an affordable price.
|
||